|
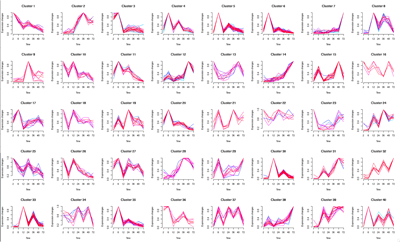
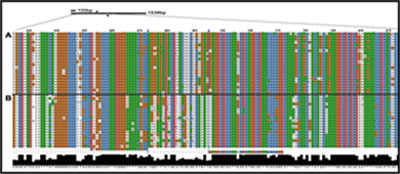
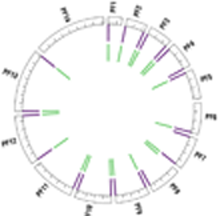
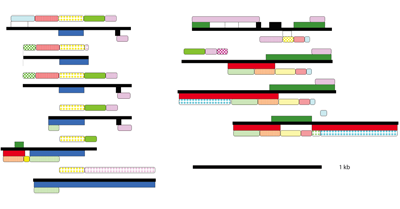
Half of the research group is dedicated to the Eukaryotic Pathogen, Vector & Host Informatics Resources Center, VEuPathDB.org (Figure E). We, WS Annotations Group, also work on the development of ontology terms and web service to enhance parasitology research and the automation of work flows, for example by making Galaxy Web Service aware (Figure F).

|